Estimating Multivariate Models with brms
Paul Bürkner
2024-03-19
Source:vignettes/brms_multivariate.Rmd
brms_multivariate.RmdIntroduction
In the present vignette, we want to discuss how to specify
multivariate multilevel models using brms. We call a
model multivariate if it contains multiple response variables,
each being predicted by its own set of predictors. Consider an example
from biology. Hadfield, Nutall, Osorio, and Owens (2007) analyzed data
of the Eurasian blue tit (https://en.wikipedia.org/wiki/Eurasian_blue_tit). They
predicted the tarsus length as well as the
back color of chicks. Half of the brood were put into
another fosternest, while the other half stayed in the
fosternest of their own dam. This allows to separate
genetic from environmental factors. Additionally, we have information
about the hatchdate and sex of the chicks (the
latter being known for 94% of the animals).
tarsus back animal dam fosternest hatchdate sex
1 -1.89229718 1.1464212 R187142 R187557 F2102 -0.6874021 Fem
2 1.13610981 -0.7596521 R187154 R187559 F1902 -0.6874021 Male
3 0.98468946 0.1449373 R187341 R187568 A602 -0.4279814 Male
4 0.37900806 0.2555847 R046169 R187518 A1302 -1.4656641 Male
5 -0.07525299 -0.3006992 R046161 R187528 A2602 -1.4656641 Fem
6 -1.13519543 1.5577219 R187409 R187945 C2302 0.3502805 FemBasic Multivariate Models
We begin with a relatively simple multivariate normal model.
bform1 <-
bf(mvbind(tarsus, back) ~ sex + hatchdate + (1|p|fosternest) + (1|q|dam)) +
set_rescor(TRUE)
fit1 <- brm(bform1, data = BTdata, chains = 2, cores = 2)As can be seen in the model code, we have used mvbind
notation to tell brms that both tarsus and
back are separate response variables. The term
(1|p|fosternest) indicates a varying intercept over
fosternest. By writing |p| in between we
indicate that all varying effects of fosternest should be
modeled as correlated. This makes sense since we actually have two model
parts, one for tarsus and one for back. The
indicator p is arbitrary and can be replaced by other
symbols that comes into your mind (for details about the multilevel
syntax of brms, see help("brmsformula")
and vignette("brms_multilevel")). Similarly, the term
(1|q|dam) indicates correlated varying effects of the
genetic mother of the chicks. Alternatively, we could have also modeled
the genetic similarities through pedigrees and corresponding relatedness
matrices, but this is not the focus of this vignette (please see
vignette("brms_phylogenetics")). The model results are
readily summarized via
fit1 <- add_criterion(fit1, "loo")
summary(fit1) Family: MV(gaussian, gaussian)
Links: mu = identity; sigma = identity
mu = identity; sigma = identity
Formula: tarsus ~ sex + hatchdate + (1 | p | fosternest) + (1 | q | dam)
back ~ sex + hatchdate + (1 | p | fosternest) + (1 | q | dam)
Data: BTdata (Number of observations: 828)
Draws: 2 chains, each with iter = 2000; warmup = 1000; thin = 1;
total post-warmup draws = 2000
Multilevel Hyperparameters:
~dam (Number of levels: 106)
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS
sd(tarsus_Intercept) 0.48 0.05 0.39 0.59 1.01 777
sd(back_Intercept) 0.24 0.07 0.11 0.39 1.00 370
cor(tarsus_Intercept,back_Intercept) -0.53 0.23 -0.94 -0.08 1.01 386
Tail_ESS
sd(tarsus_Intercept) 1155
sd(back_Intercept) 696
cor(tarsus_Intercept,back_Intercept) 410
~fosternest (Number of levels: 104)
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS
sd(tarsus_Intercept) 0.27 0.05 0.17 0.38 1.00 734
sd(back_Intercept) 0.35 0.06 0.23 0.47 1.01 335
cor(tarsus_Intercept,back_Intercept) 0.69 0.20 0.24 0.98 1.03 164
Tail_ESS
sd(tarsus_Intercept) 1199
sd(back_Intercept) 643
cor(tarsus_Intercept,back_Intercept) 514
Regression Coefficients:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
tarsus_Intercept -0.40 0.07 -0.54 -0.26 1.00 728 808
back_Intercept -0.01 0.07 -0.14 0.11 1.00 1378 1278
tarsus_sexMale 0.77 0.06 0.66 0.87 1.00 2995 1484
tarsus_sexUNK 0.23 0.13 -0.02 0.47 1.00 2672 1449
tarsus_hatchdate -0.04 0.06 -0.16 0.07 1.00 985 1308
back_sexMale 0.01 0.07 -0.12 0.14 1.00 2927 1610
back_sexUNK 0.15 0.15 -0.15 0.45 1.00 2283 1317
back_hatchdate -0.09 0.05 -0.19 0.01 1.00 1377 1436
Further Distributional Parameters:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
sigma_tarsus 0.76 0.02 0.72 0.80 1.00 1440 1334
sigma_back 0.90 0.02 0.86 0.95 1.00 2191 1511
Residual Correlations:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
rescor(tarsus,back) -0.05 0.04 -0.13 0.02 1.00 2235 1413
Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
and Tail_ESS are effective sample size measures, and Rhat is the potential
scale reduction factor on split chains (at convergence, Rhat = 1).The summary output of multivariate models closely resembles those of
univariate models, except that the parameters now have the corresponding
response variable as prefix. Within dams, tarsus length and back color
seem to be negatively correlated, while within fosternests the opposite
is true. This indicates differential effects of genetic and
environmental factors on these two characteristics. Further, the small
residual correlation rescor(tarsus, back) on the bottom of
the output indicates that there is little unmodeled dependency between
tarsus length and back color. Although not necessary at this point, we
have already computed and stored the LOO information criterion of
fit1, which we will use for model comparisons. Next, let’s
take a look at some posterior-predictive checks, which give us a first
impression of the model fit.
pp_check(fit1, resp = "tarsus")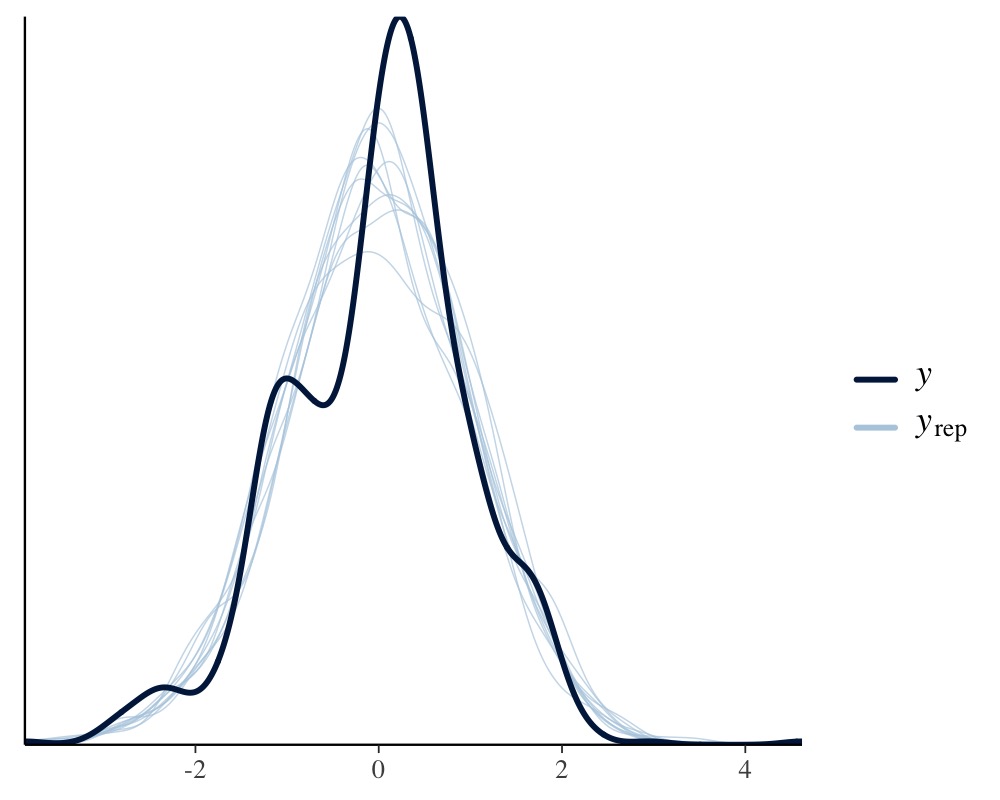
pp_check(fit1, resp = "back")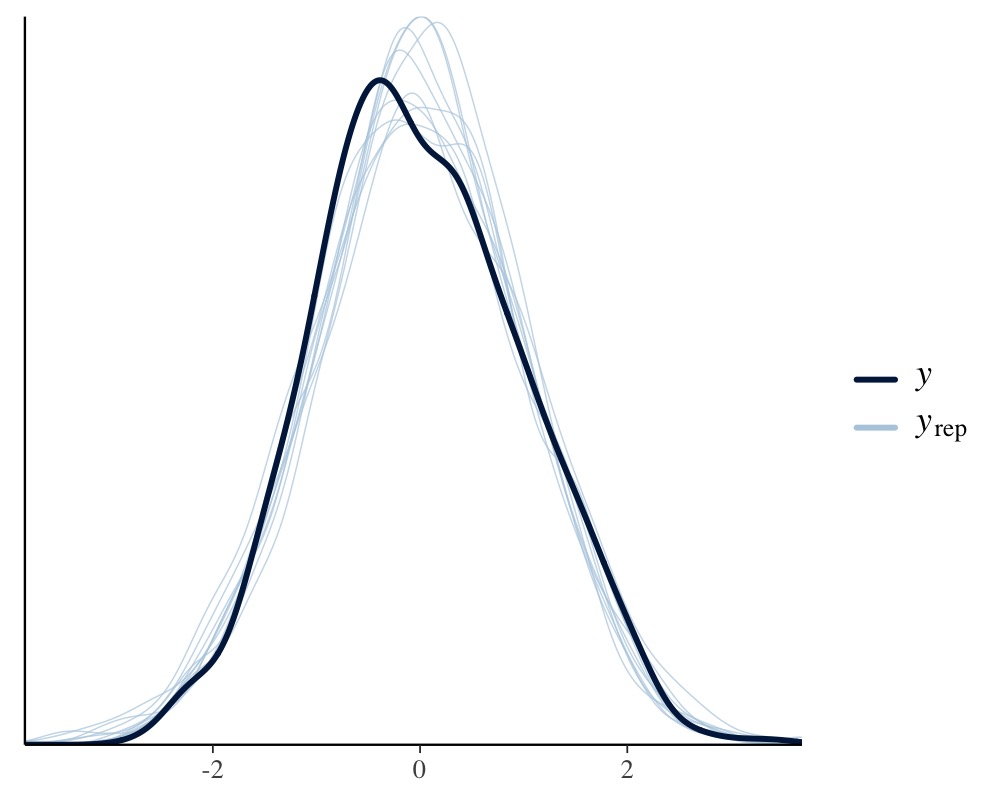
This looks pretty solid, but we notice a slight unmodeled left
skewness in the distribution of tarsus. We will come back
to this later on. Next, we want to investigate how much variation in the
response variables can be explained by our model and we use a Bayesian
generalization of the \(R^2\)
coefficient.
bayes_R2(fit1) Estimate Est.Error Q2.5 Q97.5
R2tarsus 0.4347008 0.02329837 0.3867938 0.4776906
R2back 0.1975604 0.02815540 0.1423280 0.2519281Clearly, there is much variation in both animal characteristics that we can not explain, but apparently we can explain more of the variation in tarsus length than in back color.
More Complex Multivariate Models
Now, suppose we only want to control for sex in
tarsus but not in back and vice versa for
hatchdate. Not that this is particular reasonable for the
present example, but it allows us to illustrate how to specify different
formulas for different response variables. We can no longer use
mvbind syntax and so we have to use a more verbose
approach:
bf_tarsus <- bf(tarsus ~ sex + (1|p|fosternest) + (1|q|dam))
bf_back <- bf(back ~ hatchdate + (1|p|fosternest) + (1|q|dam))
fit2 <- brm(bf_tarsus + bf_back + set_rescor(TRUE),
data = BTdata, chains = 2, cores = 2)Note that we have literally added the two model parts via
the + operator, which is in this case equivalent to writing
mvbf(bf_tarsus, bf_back). See
help("brmsformula") and help("mvbrmsformula")
for more details about this syntax. Again, we summarize the model
first.
fit2 <- add_criterion(fit2, "loo")
summary(fit2) Family: MV(gaussian, gaussian)
Links: mu = identity; sigma = identity
mu = identity; sigma = identity
Formula: tarsus ~ sex + (1 | p | fosternest) + (1 | q | dam)
back ~ hatchdate + (1 | p | fosternest) + (1 | q | dam)
Data: BTdata (Number of observations: 828)
Draws: 2 chains, each with iter = 2000; warmup = 1000; thin = 1;
total post-warmup draws = 2000
Multilevel Hyperparameters:
~dam (Number of levels: 106)
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS
sd(tarsus_Intercept) 0.48 0.05 0.39 0.58 1.00 845
sd(back_Intercept) 0.25 0.08 0.09 0.39 1.01 389
cor(tarsus_Intercept,back_Intercept) -0.49 0.22 -0.93 -0.05 1.00 580
Tail_ESS
sd(tarsus_Intercept) 1360
sd(back_Intercept) 775
cor(tarsus_Intercept,back_Intercept) 577
~fosternest (Number of levels: 104)
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS
sd(tarsus_Intercept) 0.27 0.05 0.16 0.37 1.00 666
sd(back_Intercept) 0.35 0.06 0.23 0.46 1.00 579
cor(tarsus_Intercept,back_Intercept) 0.67 0.20 0.22 0.98 1.00 316
Tail_ESS
sd(tarsus_Intercept) 1087
sd(back_Intercept) 939
cor(tarsus_Intercept,back_Intercept) 729
Regression Coefficients:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
tarsus_Intercept -0.41 0.07 -0.55 -0.28 1.00 1979 1685
back_Intercept -0.00 0.05 -0.11 0.11 1.00 2276 1509
tarsus_sexMale 0.77 0.06 0.66 0.88 1.00 3455 1538
tarsus_sexUNK 0.22 0.13 -0.03 0.47 1.00 3546 1488
back_hatchdate -0.08 0.05 -0.18 0.02 1.00 2491 1723
Further Distributional Parameters:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
sigma_tarsus 0.76 0.02 0.72 0.80 1.00 2590 1441
sigma_back 0.90 0.02 0.85 0.95 1.00 2261 1530
Residual Correlations:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
rescor(tarsus,back) -0.05 0.04 -0.13 0.02 1.00 2498 1746
Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
and Tail_ESS are effective sample size measures, and Rhat is the potential
scale reduction factor on split chains (at convergence, Rhat = 1).Let’s find out, how model fit changed due to excluding certain effects from the initial model:
loo(fit1, fit2)Output of model 'fit1':
Computed from 2000 by 828 log-likelihood matrix.
Estimate SE
elpd_loo -2126.9 33.6
p_loo 176.0 7.4
looic 4253.8 67.3
------
MCSE of elpd_loo is NA.
MCSE and ESS estimates assume MCMC draws (r_eff in [0.4, 1.9]).
Pareto k diagnostic values:
Count Pct. Min. ESS
(-Inf, 0.7] (good) 827 99.9% 85
(0.7, 1] (bad) 1 0.1% <NA>
(1, Inf) (very bad) 0 0.0% <NA>
See help('pareto-k-diagnostic') for details.
Output of model 'fit2':
Computed from 2000 by 828 log-likelihood matrix.
Estimate SE
elpd_loo -2126.0 33.9
p_loo 176.1 7.8
looic 4252.0 67.8
------
MCSE of elpd_loo is NA.
MCSE and ESS estimates assume MCMC draws (r_eff in [0.5, 1.7]).
Pareto k diagnostic values:
Count Pct. Min. ESS
(-Inf, 0.7] (good) 826 99.8% 79
(0.7, 1] (bad) 2 0.2% <NA>
(1, Inf) (very bad) 0 0.0% <NA>
See help('pareto-k-diagnostic') for details.
Model comparisons:
elpd_diff se_diff
fit2 0.0 0.0
fit1 -0.9 1.3 Apparently, there is no noteworthy difference in the model fit.
Accordingly, we do not really need to model sex and
hatchdate for both response variables, but there is also no
harm in including them (so I would probably just include them).
To give you a glimpse of the capabilities of brms’
multivariate syntax, we change our model in various directions at the
same time. Remember the slight left skewness of tarsus,
which we will now model by using the skew_normal family
instead of the gaussian family. Since we do not have a
multivariate normal (or student-t) model, anymore, estimating residual
correlations is no longer possible. We make this explicit using the
set_rescor function. Further, we investigate if the
relationship of back and hatchdate is really
linear as previously assumed by fitting a non-linear spline of
hatchdate. On top of it, we model separate residual
variances of tarsus for male and female chicks.
bf_tarsus <- bf(tarsus ~ sex + (1|p|fosternest) + (1|q|dam)) +
lf(sigma ~ 0 + sex) + skew_normal()
bf_back <- bf(back ~ s(hatchdate) + (1|p|fosternest) + (1|q|dam)) +
gaussian()
fit3 <- brm(
bf_tarsus + bf_back + set_rescor(FALSE),
data = BTdata, chains = 2, cores = 2,
control = list(adapt_delta = 0.95)
)Again, we summarize the model and look at some posterior-predictive checks.
fit3 <- add_criterion(fit3, "loo")
summary(fit3) Family: MV(skew_normal, gaussian)
Links: mu = identity; sigma = log; alpha = identity
mu = identity; sigma = identity
Formula: tarsus ~ sex + (1 | p | fosternest) + (1 | q | dam)
sigma ~ 0 + sex
back ~ s(hatchdate) + (1 | p | fosternest) + (1 | q | dam)
Data: BTdata (Number of observations: 828)
Draws: 2 chains, each with iter = 2000; warmup = 1000; thin = 1;
total post-warmup draws = 2000
Smoothing Spline Hyperparameters:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
sds(back_shatchdate_1) 1.97 0.99 0.28 4.35 1.00 580 564
Multilevel Hyperparameters:
~dam (Number of levels: 106)
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS
sd(tarsus_Intercept) 0.47 0.05 0.38 0.57 1.00 905
sd(back_Intercept) 0.24 0.07 0.10 0.37 1.01 373
cor(tarsus_Intercept,back_Intercept) -0.52 0.22 -0.95 -0.06 1.00 699
Tail_ESS
sd(tarsus_Intercept) 1232
sd(back_Intercept) 778
cor(tarsus_Intercept,back_Intercept) 812
~fosternest (Number of levels: 104)
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS
sd(tarsus_Intercept) 0.26 0.05 0.16 0.37 1.00 597
sd(back_Intercept) 0.31 0.06 0.20 0.44 1.01 565
cor(tarsus_Intercept,back_Intercept) 0.64 0.22 0.13 0.97 1.00 292
Tail_ESS
sd(tarsus_Intercept) 1030
sd(back_Intercept) 1088
cor(tarsus_Intercept,back_Intercept) 758
Regression Coefficients:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
tarsus_Intercept -0.41 0.07 -0.54 -0.28 1.00 1462 1679
back_Intercept 0.00 0.05 -0.10 0.10 1.00 2388 1720
tarsus_sexMale 0.77 0.06 0.66 0.88 1.00 4603 1343
tarsus_sexUNK 0.22 0.12 -0.01 0.45 1.00 3206 1487
sigma_tarsus_sexFem -0.30 0.04 -0.38 -0.22 1.00 3599 1612
sigma_tarsus_sexMale -0.25 0.04 -0.33 -0.16 1.00 3720 1388
sigma_tarsus_sexUNK -0.39 0.13 -0.63 -0.14 1.00 3120 1537
back_shatchdate_1 -0.27 3.18 -6.00 7.05 1.00 1163 891
Further Distributional Parameters:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
sigma_back 0.90 0.02 0.86 0.95 1.00 2467 1611
alpha_tarsus -1.25 0.38 -1.86 -0.31 1.00 2128 1025
Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
and Tail_ESS are effective sample size measures, and Rhat is the potential
scale reduction factor on split chains (at convergence, Rhat = 1).We see that the (log) residual standard deviation of
tarsus is somewhat larger for chicks whose sex could not be
identified as compared to male or female chicks. Further, we see from
the negative alpha (skewness) parameter of
tarsus that the residuals are indeed slightly left-skewed.
Lastly, running
conditional_effects(fit3, "hatchdate", resp = "back")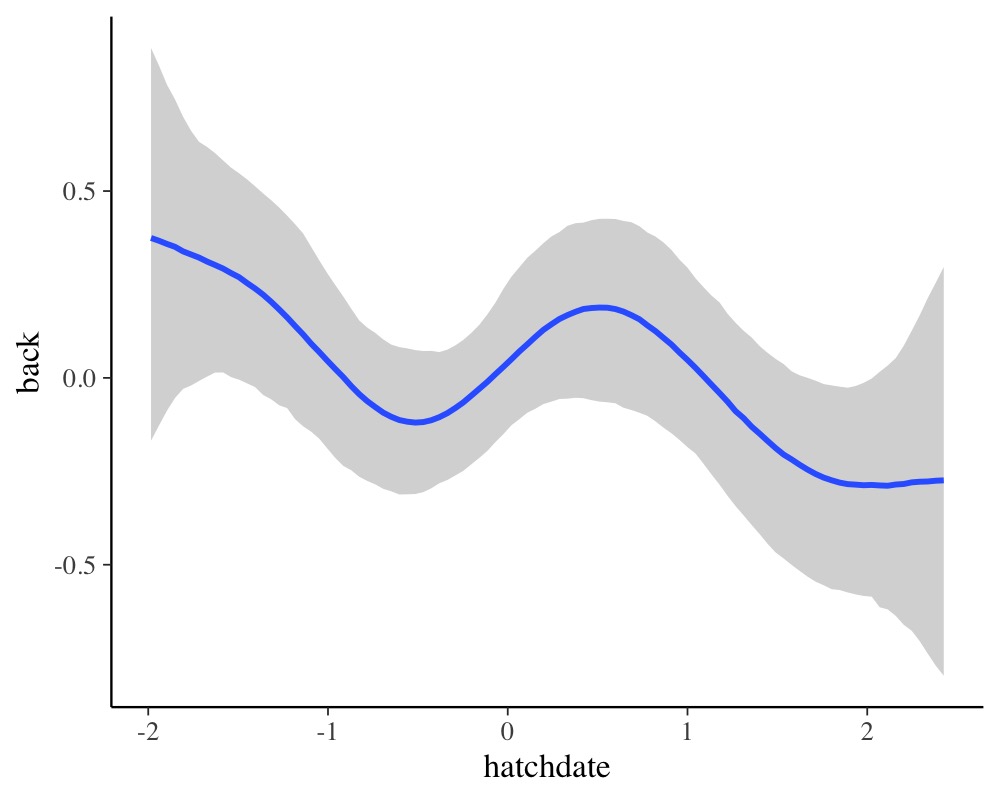
reveals a non-linear relationship of hatchdate on the
back color, which seems to change in waves over the course
of the hatch dates.
There are many more modeling options for multivariate models, which
are not discussed in this vignette. Examples include autocorrelation
structures, Gaussian processes, or explicit non-linear predictors (e.g.,
see help("brmsformula") or
vignette("brms_multilevel")). In fact, nearly all the
flexibility of univariate models is retained in multivariate models.